March 27, 2024
Data-structures for sets of sequences: a third update
This post starts with the exploration initiated in an earlier paper, and blog post, and today I’ll present the fresh contributions of 2023-2024. Inspired by a moment of reflection shared on by Lior Pachter about his blog, I’ve decided to incorporate DOIs in my posts so they can be cited.
For those diving into this post, a prerequisite is the understanding of sequence bioinformatics concepts, such as the BWT, minimizers, de Bruijn graphs (you can start with the previous citations).
Today’s discussion revolves around the following figure. I adopted a solution that lays out data structures on twin discs, because honeslty, a standard tree or a table takes just too much space to be correctly organised. The left disc shows structures enabling k-mer set representation. Representation is a vague word, I make an attempt at categorizing what the structures actually do using colors. I utilize several disc radius to illustrate how fundamental concepts such as hashing or bit vectors are derived and associated with useful tools (eg minimizers) to obtain the final structures, that appear on the outermost radius.
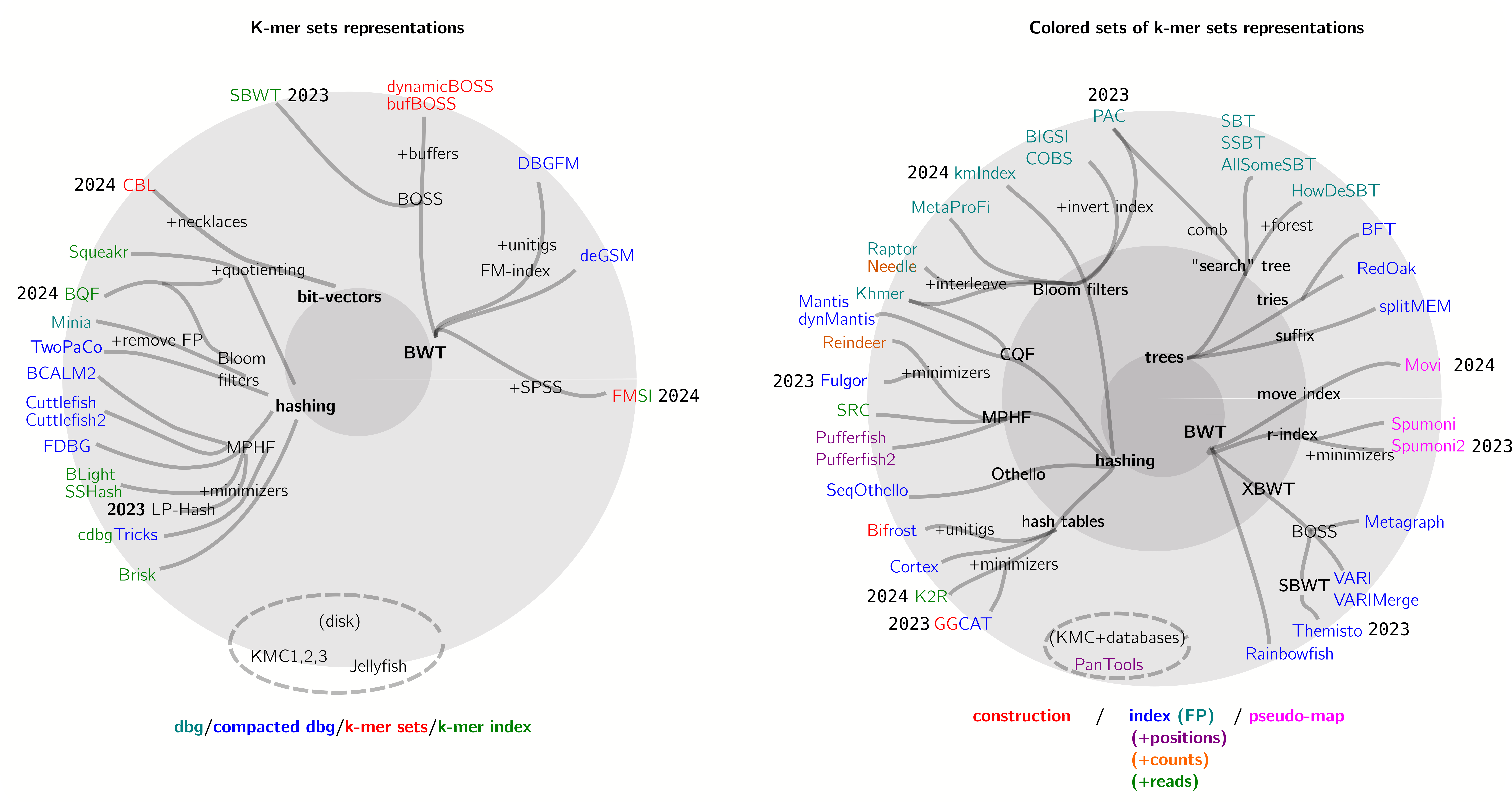
In the right disc, I show colored k-mer set collections. “Colored” in this context means each k-mer set bears metadata, typically denoting its dataset of origin (it is totally different from the colors I use in the legend). It quite natural that colored k-mer set collections integrate structures from the left disc, hence some of the methods in the inner circles of the right disc already appear in the left one.
Each disc comes with a color legend, to detail features:
-
Left disc legends denote (dbg) structures facilitating navigation within a de Bruijn graph, (compacted bdg) assembling unitigs from such a graph, (k-mer set) enabling operations on a k-mer set (all structures support membership), and (k-mer index) constructing a k-mer index for mapping k-mers, possibly with specific attributes like abundance.
-
Right disc annotations focus on (construction)methods yielding unitigs in FASTA format, tagged with their color information, and (indexing) is methods mapping k-mers to their origins (with possible false positives) and/or sometimes additional metadata retention such as positions in the initial sequences, counts in each source dataset, and read occurrences, and (pseudo-map) means that sequences are matched to regions of the set of sequences.
Insights
There are, in fact, many ways to categorize the structures, this is one attempt. The disc organization revolves around filiation between data structures, while the colors reflect more end user experience on features. But features are numerous. For instance, the capacity to extract other superstrings of k-mer of interest, such as simplitigs or eulertigs, is not represented, but for instance possible in GGCAT.
Similarly, the filiation can be discussed. Many contributions are a patchwork of ideas. Some are hybrids (eg PAC or BQF). Navigating the BWT landscape is difficult (at least for me), and I hope it will be the next topic of a future post. For instance here, I omit relatedness to Wheeler graphs. It is noteworthy that some of the presented indexes based on the BWT can represent sequences longer than the k-mers, that is the case for MOVI and SPUMONI 1 and 2 (other structures are specialized towards k-mers).
Finally, note that some concepts are disseminated in so many works that I could not totally factorize them, this is especially the case for minimizers.
Detailing the left disc:
- Squeakr and the more recent BQF are designed for k-mer abundance association (also called Counting Quotient filters). They can be exact, but perform in their optimal setting if some count overestimation can be allowed. BQF lowers this false positive rate for a similar space requirement.
- LP-hash introduces a cache- and memory-efficient hashing approach for k-mers, pertaining to the minimal perfect hashing (MPHF) family. It is not yet used in a k-mer index.
- Two recent contributions, CBL and FMSI allow set operations such as intersection, union and differences on k-mer sets, these features were strikingly missing in the previous structures.
- People with FOMO will look everywhere about cdgbTricks and Brisk, these are works in progress that I expect to come out in 2024.
On the right disc:
- Bifrost can be considered an older tool than many of the presented one, and is still considered a reference. In my personal opinion, it can be explained not because it is the best at a single performance metric, or at having a striking different feature, but because it was designed to be maintained on the long term, and has be usable since the beginning. It therefore amassed a large and engaged community of users.
- Minimizers and spectrum preserving string sets have been ingested in many hash-based tools.
- Many tools have focused on cache efficiency by ensuring that groups of consecutive k-mers are processed together in the cache. Other methods owe their high efficiency to memory mapping (kmindex).
- BWT-based methods have recently advanced by integrating move index operations, enabling benefits from both the space complexity of O(r), with r being the number of runs in the initial BWT of the data, and from a O(1) query time, which used to be a limiting factor (e.g., in MOVI).
Global notes:
-
Colored k-mer sets are recently used for pangenome annotation here and here and for metagenomic profiling with KMCP. Will 2024 be the year of applications of these methods? Metagraph’s team was a pioneer at releasing online large scale indexes to explore collections of datasets, but in 2024 we’ve seen at least 3 new manuscripts. ORA (manuscript/website) allows to navigate the sequencing of plankton) all over the globe, along with biotic and abiotic factors. AllTheBacteria (manuscript) provides a huge collection of queriable assembled bacterial genomes (almost 2 million!). The Transipedia index (manuscript/website) https://kamimrcht.github.io/webpage/tigs.htmlshows how a large scale k-mer RNA-seq cancer database allows to spot cancer biomarkers.
-
3 of the mentioned 2023-2024 structures were developed in Rust, the majority of the rest in C++, some with projects of reimplementations in Rust.
-
Metadata inclusion and compression in the graphs remains an understudied topic, but with notable efforts here and here. For sequence compression, outside of the BWT approaches, the compacted representations of k-mers sets through strings leveraging k-mer overlaps (spectral preserving string sets SPSS) have a unifiying paper, and phylogenetic compression allowed to compress the 2 million assemblies in AllTheBacteria.
-
I think we will witness more and more convergence between structures described here and variation graphs. I also wish there were more connections done between the BWT paradigm and the hashing paradigm.
-
If we restrict to data structures manuscripts only (and put aside applications), how many distinct female authors can we account for in the literature cited in the figure? Let’s name them. Fatemeh Almodaresi, Amatur Rahman, Mitra Darvish, Svenja Mehringer, Bahar Alipanahi, Léa Vandamme, Olga Kalinina, Annie Chateau, Christina Boucher and myself. There is also a small amount of groups represented in my review, most being in Europe or North America. Some authors from these hubs come from lower income countries such as Bangladesh or Brasil. Other exceptions include a few chinese colleagues: Wei Shen, Hongzhe Guo and his collaborators. These observations could very well, partly at least, be a reflect of biases of my personnal scientific bubble.
References of newest papers:
Structures that can handle k-mer sets:
- CBL, dynamic, with many set operations
- FMSI, based on efficient SPSS, allowing boolean operators on sets, low memory with time efficient queries
- BQF, if storing abundances is a requirement
- LP-hash a highly efficient, static hash function for k-mer sets
Structures for colored sets:
- Fulgor currently the most space efficient for storing collections of datasets, see this post on covid pangenomes
- Themisto another solution based on the SBWT
- Movi with interesting space scalability for a growing corpus of close genomes
- Spumoni2 presents a highly compressed scheme for collections of genomes
- kmindex full disk and Bloom filters based, very scalable
- PAC allow especially quick queries in diverse collections
- K2R links k-mers to their origin reads, provides a compression for long reads sets
- GGCAT will build a colored de Bruijn graph, compacted unitigs, eulertigs and matchtigs
Edit 03/28: Simon Puglisi reminded me to add Bahar Alipanahi to the list of contributing women.